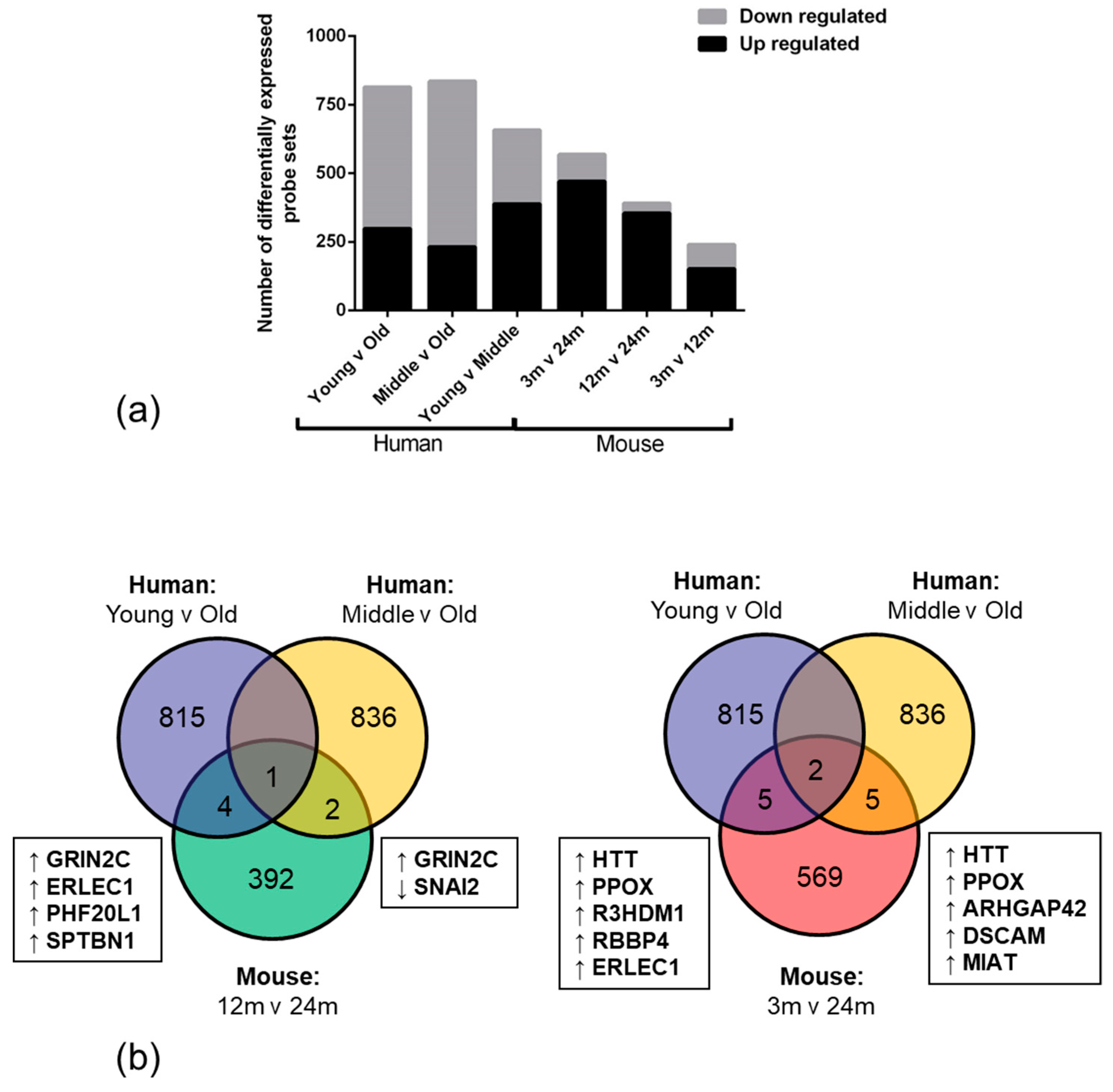
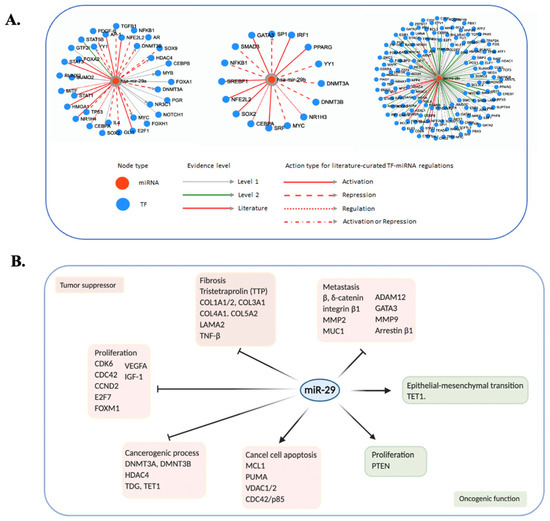
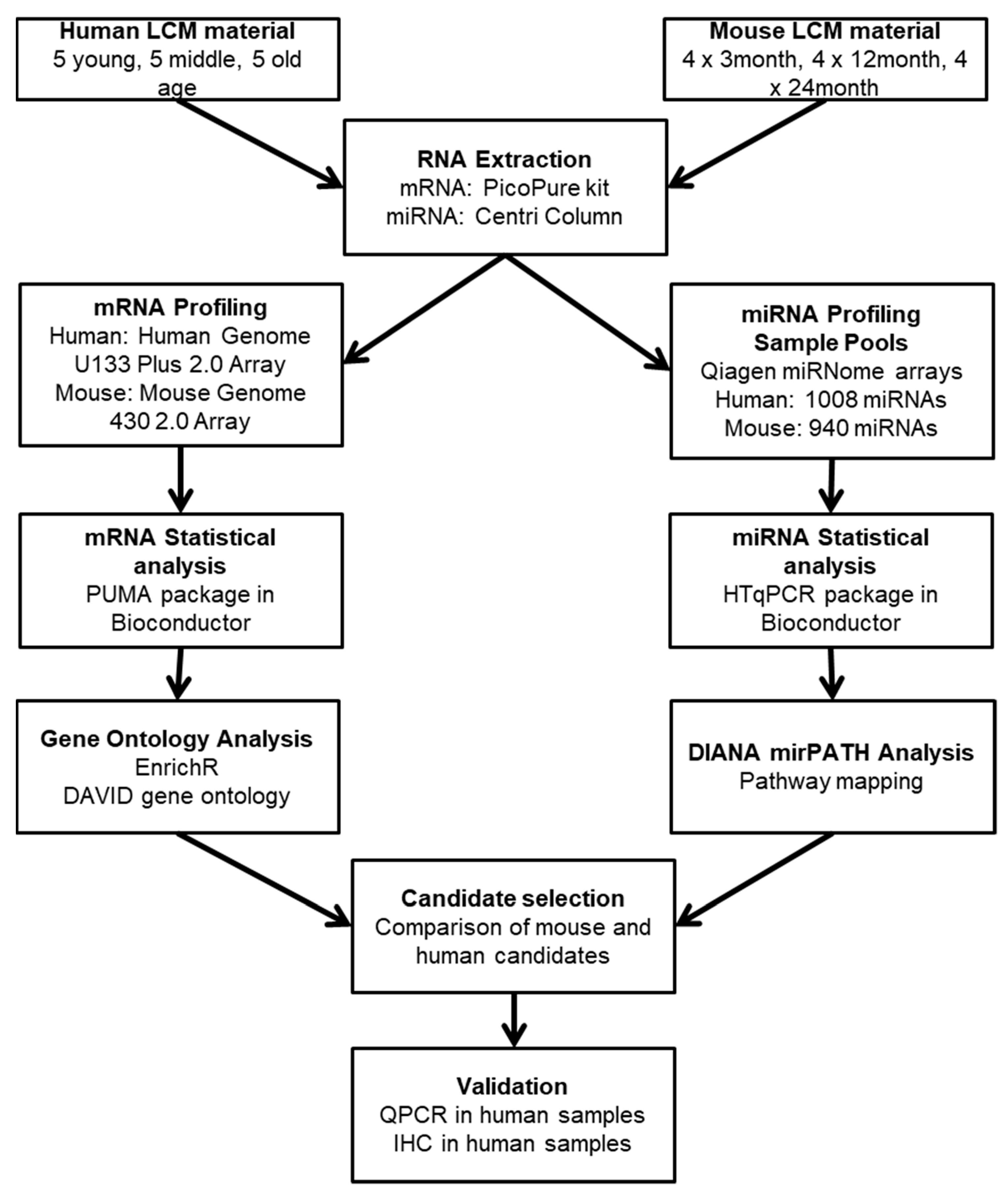
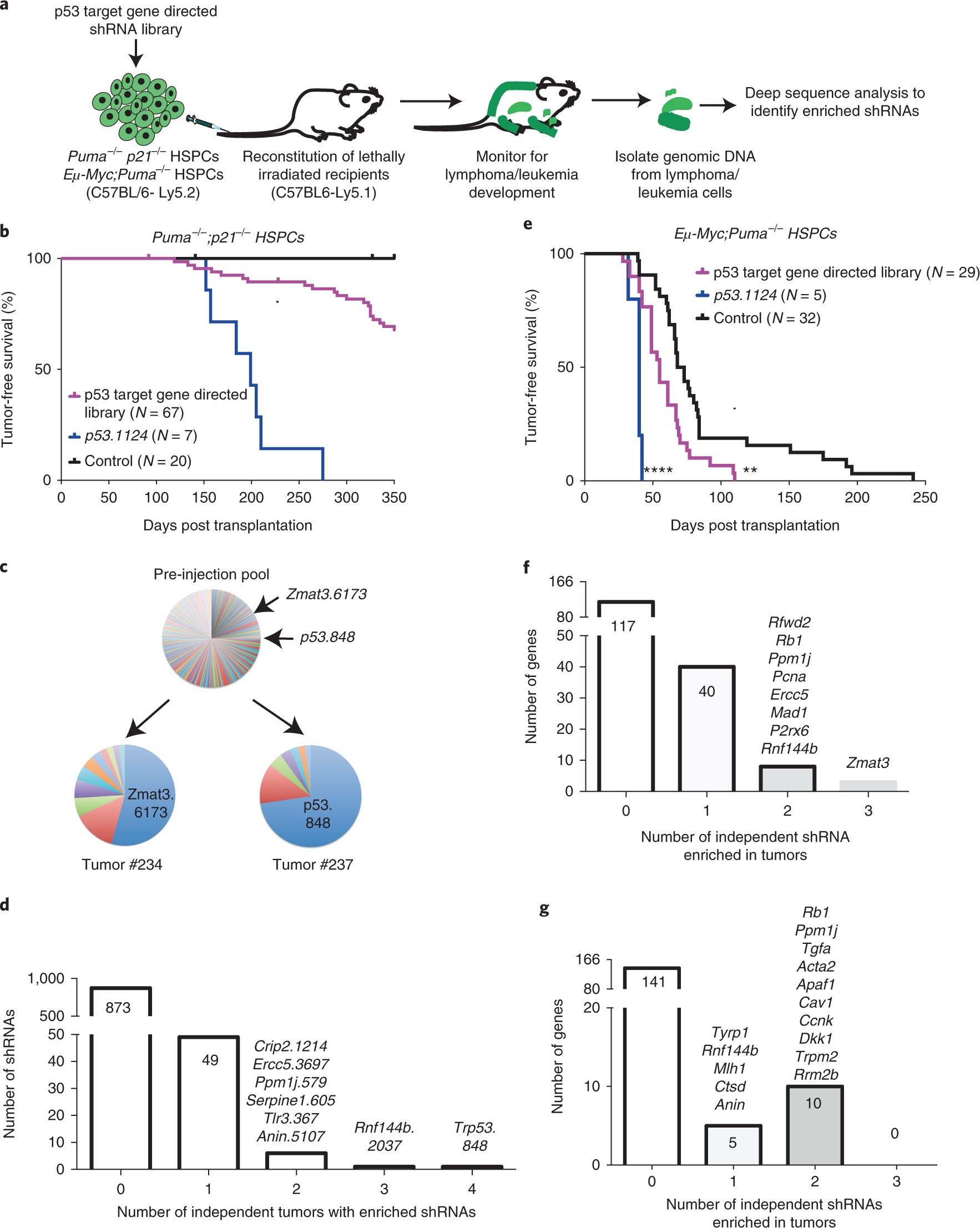
Nonsense-mediated mRNA decay inhibition synergizes with MDM2 inhibition to suppress TP53 wild-type cancer cells in p53 isoform-dependent manner | Cell Death Discovery

puma 3.0: improved uncertainty propagation methods for gene and transcript expression analysis | BMC Bioinformatics | Full Text
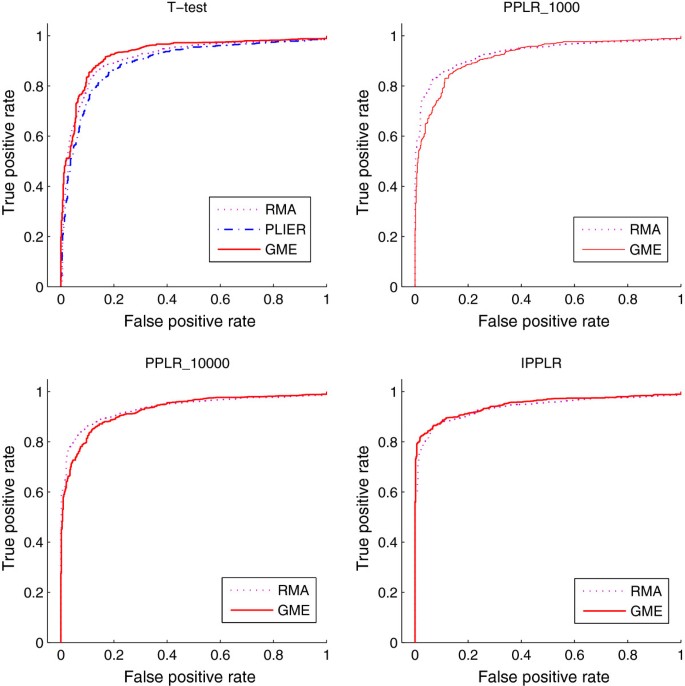
puma 3.0: improved uncertainty propagation methods for gene and transcript expression analysis | BMC Bioinformatics | Full Text
Bioinformatics-R/2.1 Installing packages from Bioconductor.md at master · xiaowei3223/Bioinformatics-R · GitHub
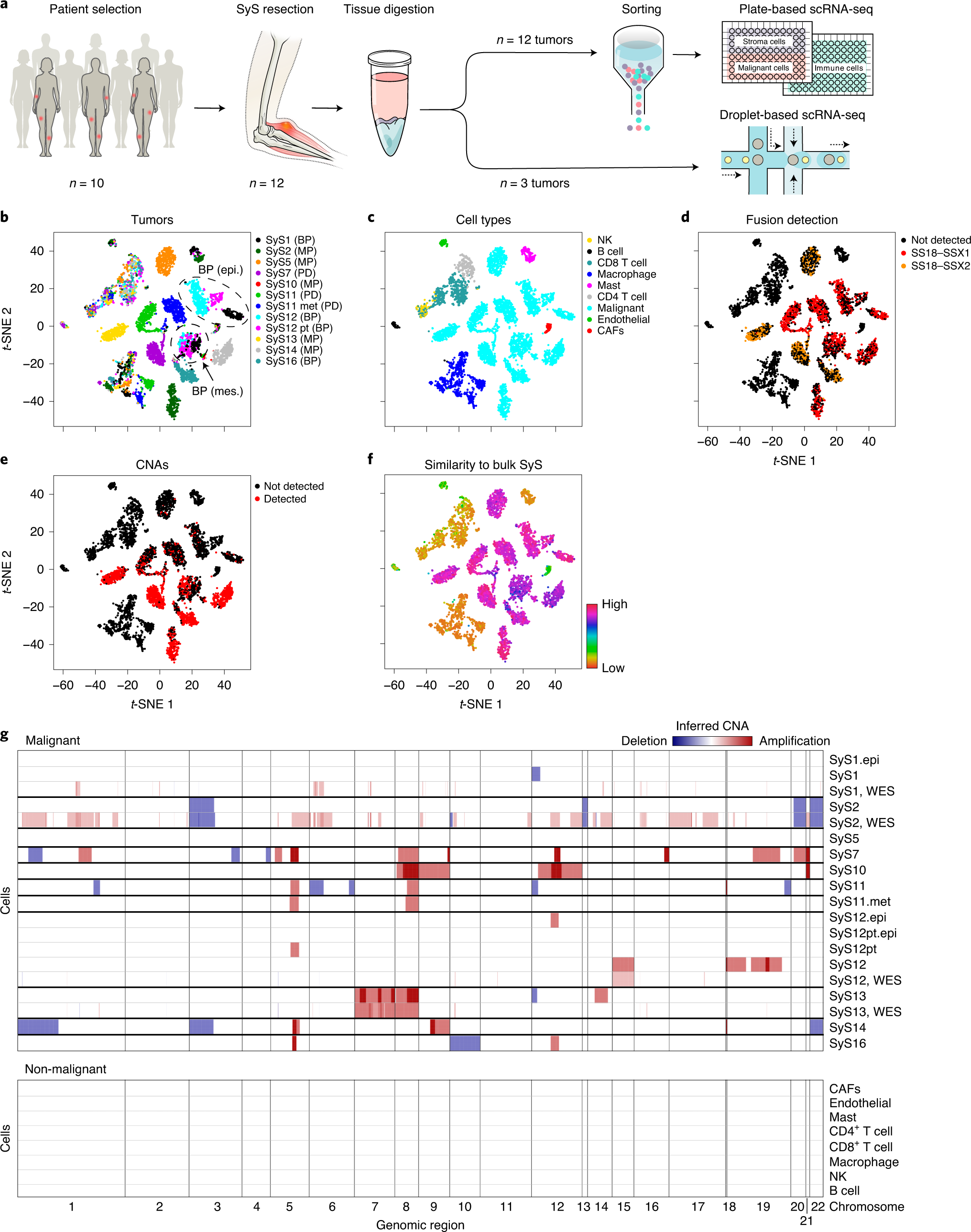
Opposing immune and genetic mechanisms shape oncogenic programs in synovial sarcoma | Nature Medicine

puma 3.0: improved uncertainty propagation methods for gene and transcript expression analysis | BMC Bioinformatics | Full Text
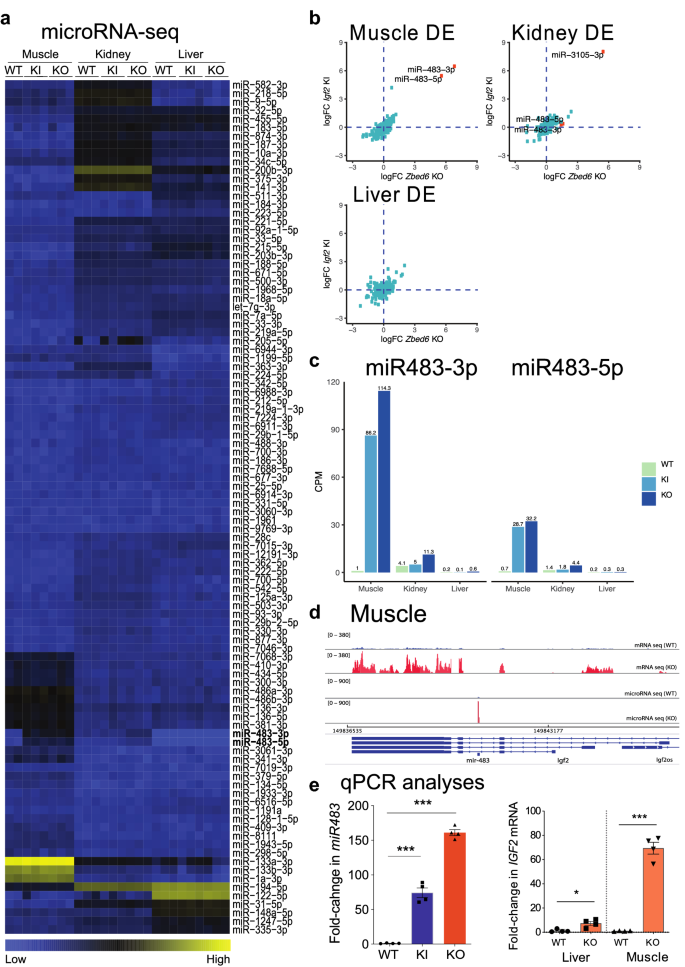
ZBED6 regulates Igf2 expression partially through its regulation of miR483 expression | Scientific Reports

Comprehensive Transcriptomic Analysis of Novel Class I HDAC Proteolysis Targeting Chimeras (PROTACs) | Biochemistry
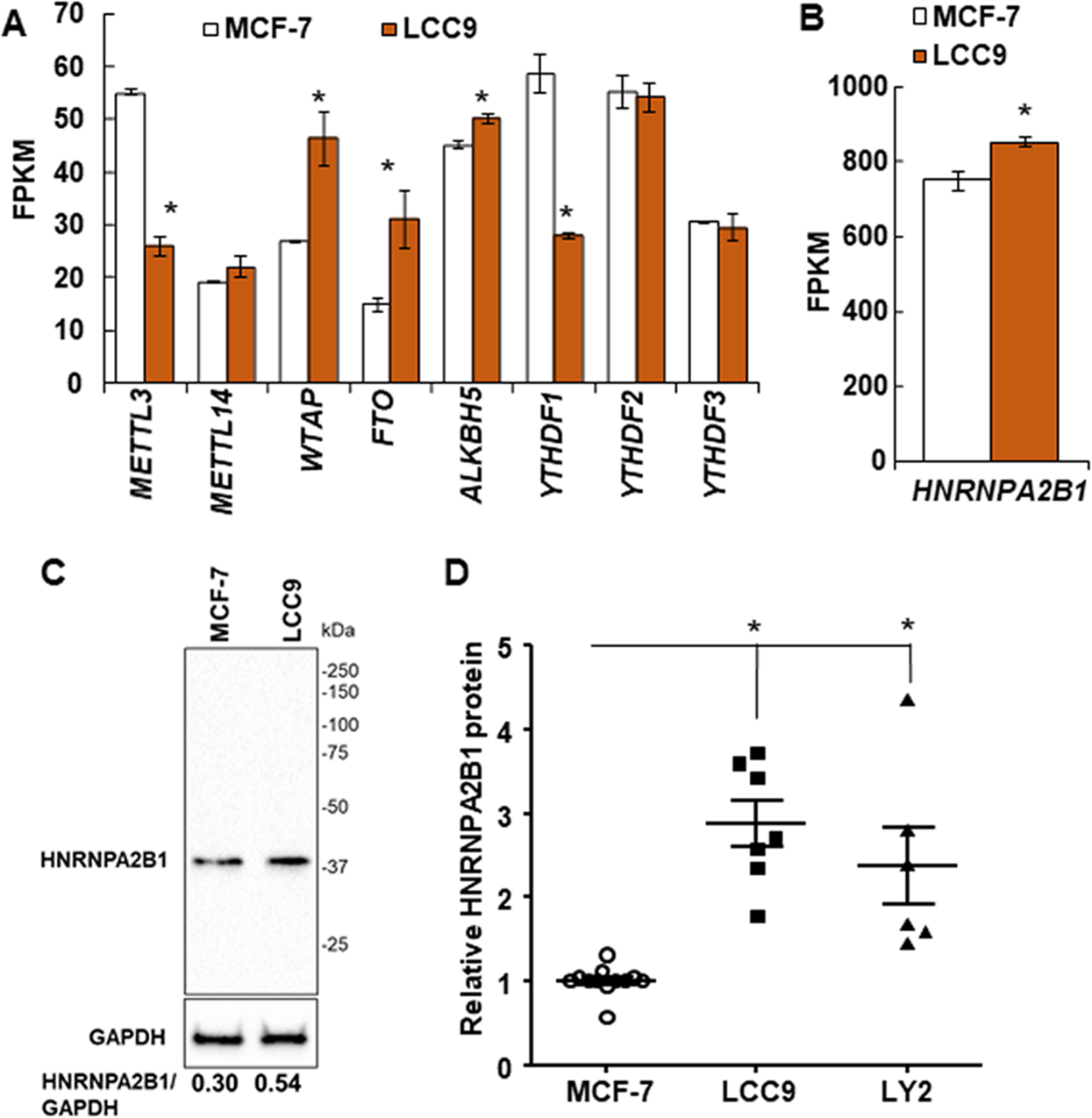
HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells | Scientific Reports
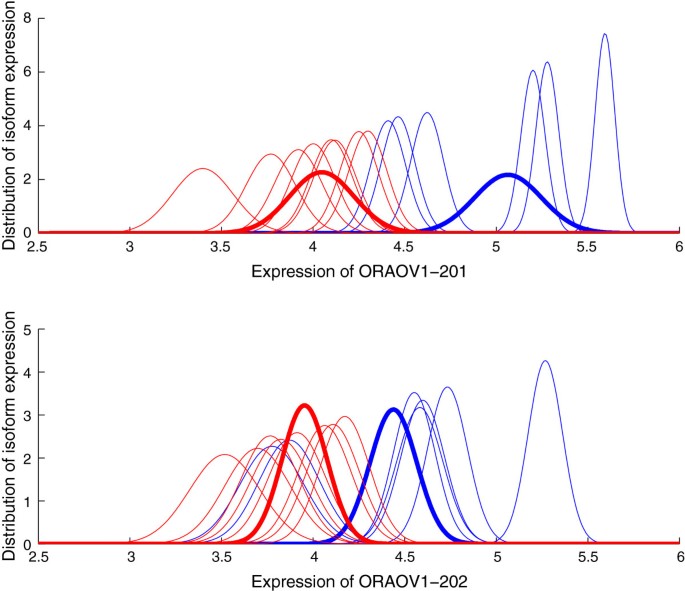
puma 3.0: improved uncertainty propagation methods for gene and transcript expression analysis | BMC Bioinformatics | Full Text

Nutlin-Induced Apoptosis Is Specified by a Translation Program Regulated by PCBP2 and DHX30 - ScienceDirect

Identification of key biomarkers for thyroid cancer by integrative gene expression profiles - Jinyi Tian, Yizhou Bai, Anyang Liu, Bin Luo, 2021

Diagnostics | Free Full-Text | Comparative Genomic Hybridization and Transcriptome Sequencing Reveal Genes with Gain in Acute Lymphoblastic Leukemia: JUP Expression Emerges as a Survival-Related Gene

Targeting mitochondrial respiration and the BCL2 family in high‐grade MYC‐associated B‐cell lymphoma - Donati - 2022 - Molecular Oncology - Wiley Online Library

Transient regulation of focal adhesion via Tensin3 is required for nascent oligodendrocyte differentiation | eLife